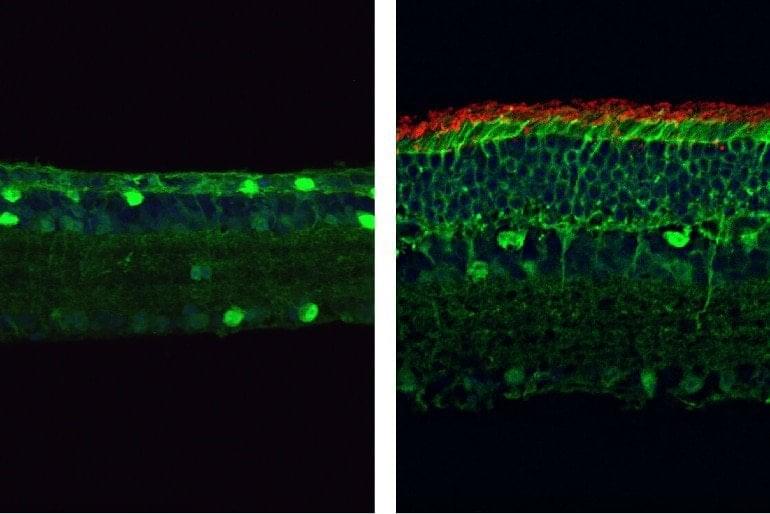
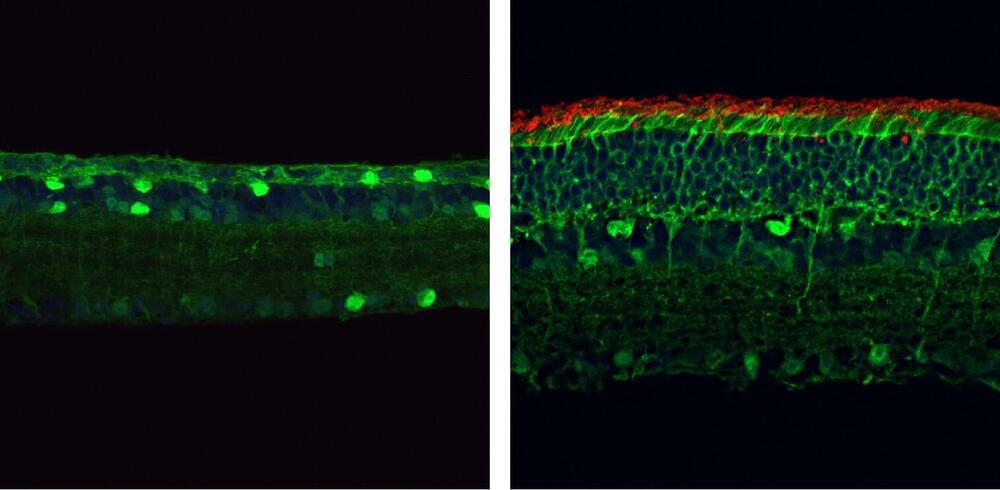
Researchers at Texas A&M University have developed the first molecular therapeutic for Angelman syndrome to advance into clinical development.
In a new article, published today in Science Translational Medicine, Dr. Scott Dindot, an associate professor and EDGES Fellow in the Texas A&M School of Veterinary Medicine and Biomedical Sciences’ (VMBS) Department of Veterinary Pathobiology, and his team share the process through which they developed this novel therapeutic candidate, also known as 4.4.PS.L, or GTX-102. Dindot is also the executive director of molecular genetics at Ultragenyx, which is leading the development of GTX-102.
Angelman syndrome (AS) is a devastating, rare neurogenetic disorder that affects approximately 1 in 15,000 live births per year; the disorder is triggered by a loss of function of the maternal UBE3A gene in the brain, causing developmental delay, absent speech, movement or balance disorder, and seizures.