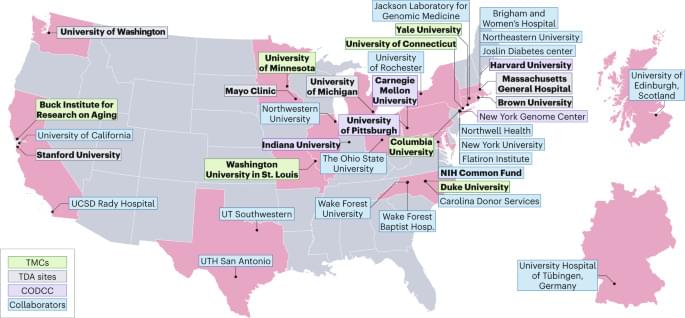
TMCs are responsible for all aspects of data generation from tissue collection and analysis to data integration and interpretation. We anticipate that TMCs will acquire and integrate imaging and omics data to benchmark, standardize and validate SnC maps at single-cell resolution for their assigned tissues. The TDA sites are responsible for development of innovative, new approaches and tools necessary to deeply phenotype SnCs in human tissues and model systems. Examples include multi-omics characterization of the 4D nucleome in SnCs, high-throughput quantification of telomere-associated foci, and in vivo detection of SnCs via positron emission tomography imaging. Once developed, these new technologies are expected to be applied broadly and collaboratively across multiple tissues by the TMCs. The CODCC will collect, store and curate all data and metadata generated by the TMCs and TDA sites. The CODCC is responsible for generating the computational models, and final atlas products as well as the tools to visualize and disseminate the data as a resource for the broad scientific community.
It is expected that SenNet will interface with other cell mapping programs such as Human Bimolecular Atlas Program (HuBMAP), Human Cell Atlas (HCA) and the Kidney Precision Medicine Project (KPMP). HuBMAP is an NIH Common Fund Initiative to develop the resources and framework to map the 30 trillion cells that make up the human body using protein identifiers of cell lineage. HCA is using single-cell and spatial transcriptomics to create cell reference maps defining the position, function and characteristics of all cells in the human body. The KPMP is an initiative of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) aimed at using state-of-the-art and emerging technologies to characterize renal biopsies from participants with acute kidney injury or chronic kidney disease to enable personalized approaches to their treatment.