Hackathons using AlphaGenome and other AI models are hunting down the genetic causes of devastating conditions that have evaded diagnosis.



Plants display a wide range of life spans and aging rates. Although dynamic changes to DNA methylation are a hallmark of aging in mammals, it is unclear whether similar molecular signatures reflect rates of aging and organism life span in plants. In this work, we show that the short-lived model plant Arabidopsis thaliana exhibits a loss of epigenetic integrity during aging, which causes DNA methylation decay and the expression of transposable elements. We show that the rate of epigenetic aging can be manipulated by extending or curtailing life span and that shoot apical meristems are protected from these epigenetic changes. We demonstrate that a program of transcriptional repression suppresses DNA methylation maintenance pathways during aging and that mutants of this program display a complete absence of epigenetic decay while physical aging remains unaffected.
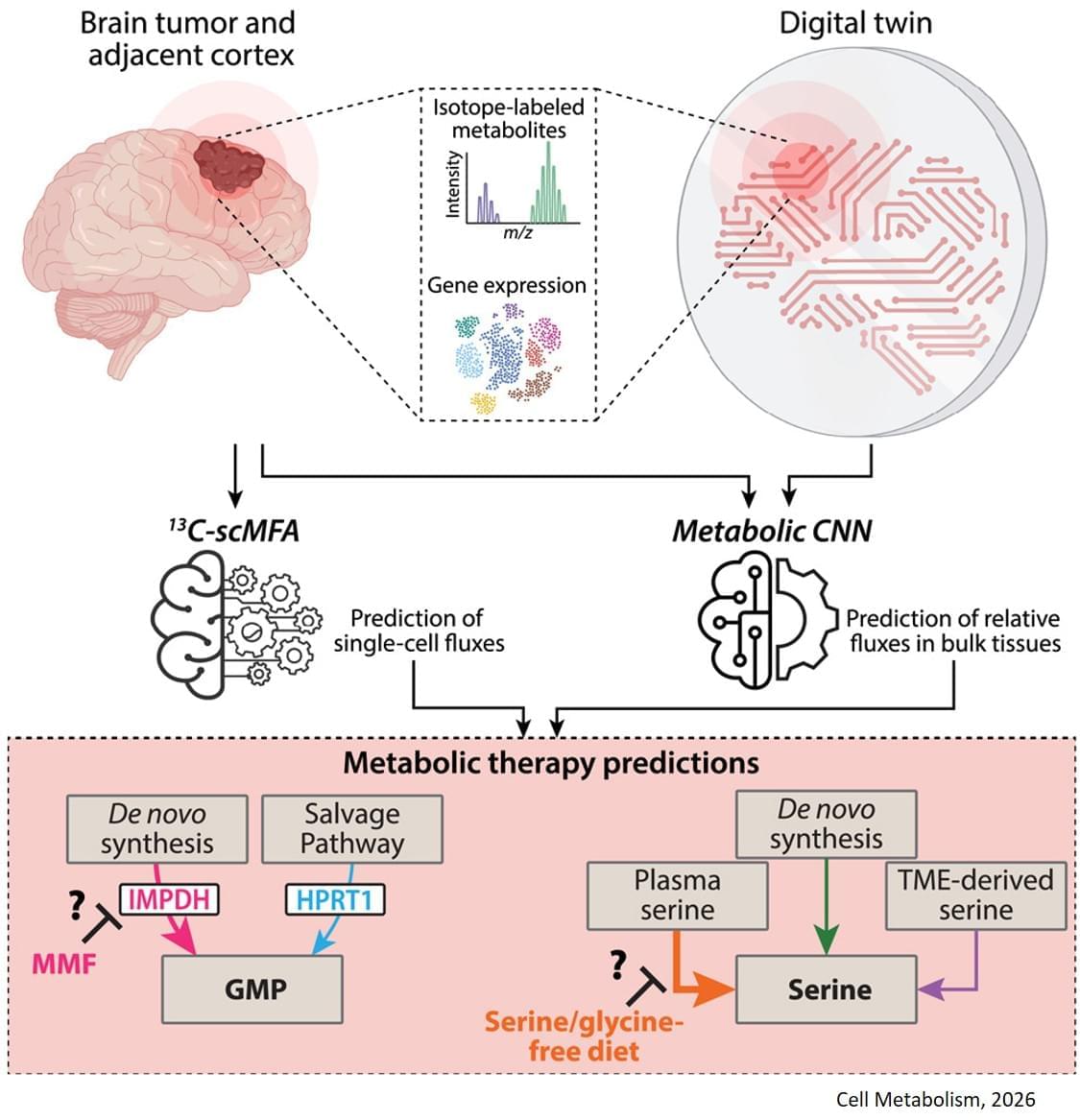
The study, published in Cell Metabolism, builds on previous research showing that some gliomas can be slowed down through the patient’s diet. If a patient isn’t consuming certain protein building blocks, called amino acids, then some tumors are unable to grow. However, other tumors can produce these amino acids for themselves, and can continue growing anyway. Until now, there was no easy way to tell which patients would benefit from dietary restrictions.
The digital twin’s ability to map metabolic activity in tumors also helped determine whether a drug that prevents tumors from producing a building block for replicating and repairing DNA would work, as some cells can obtain that molecule from their environments.
To overcome challenges in mapping tumor metabolism inside the brain, the team developed a computer-based “digital twin” that can predict how an individual patient’s brain tumor will react to each treatment.
“Typically, metabolic measurements during surgeries to remove tumors can’t provide a clear picture of tumor metabolism—surgeons can’t observe how metabolism varies with time, and labs are limited to studying tissues after surgery. By integrating limited patient data into a model based on fundamental biology, chemistry and physics, we overcame these obstacles,” said a co-corresponding author of the study.
The digital twin uses patient data obtained through blood draws, metabolic measurements of the tumor tissue and the tumor’s genetic profile. The digital twin then calculates the speed at which the cancer cells consume and process nutrients, known as metabolic flux.
“This is the first time a machine learning and AI-based approach has been used to measure metabolic flux directly in patient tumors,” said a co-first author of the study.
The researchers built a type of deep learning model called a convolutional neural network and trained it on synthetic patient data, generated based on known biology and chemistry and constrained by measurements from eight patients with glioma who were infused with labeled glucose during surgery. By comparing their computer models with different data from six of those patients, they found the digital twins could predict metabolic activity with high accuracy. In experiments conducted on mice, the team confirmed that the diet only slowed tumor growth in mice that the digital twin had identified as good candidates for the treatment.
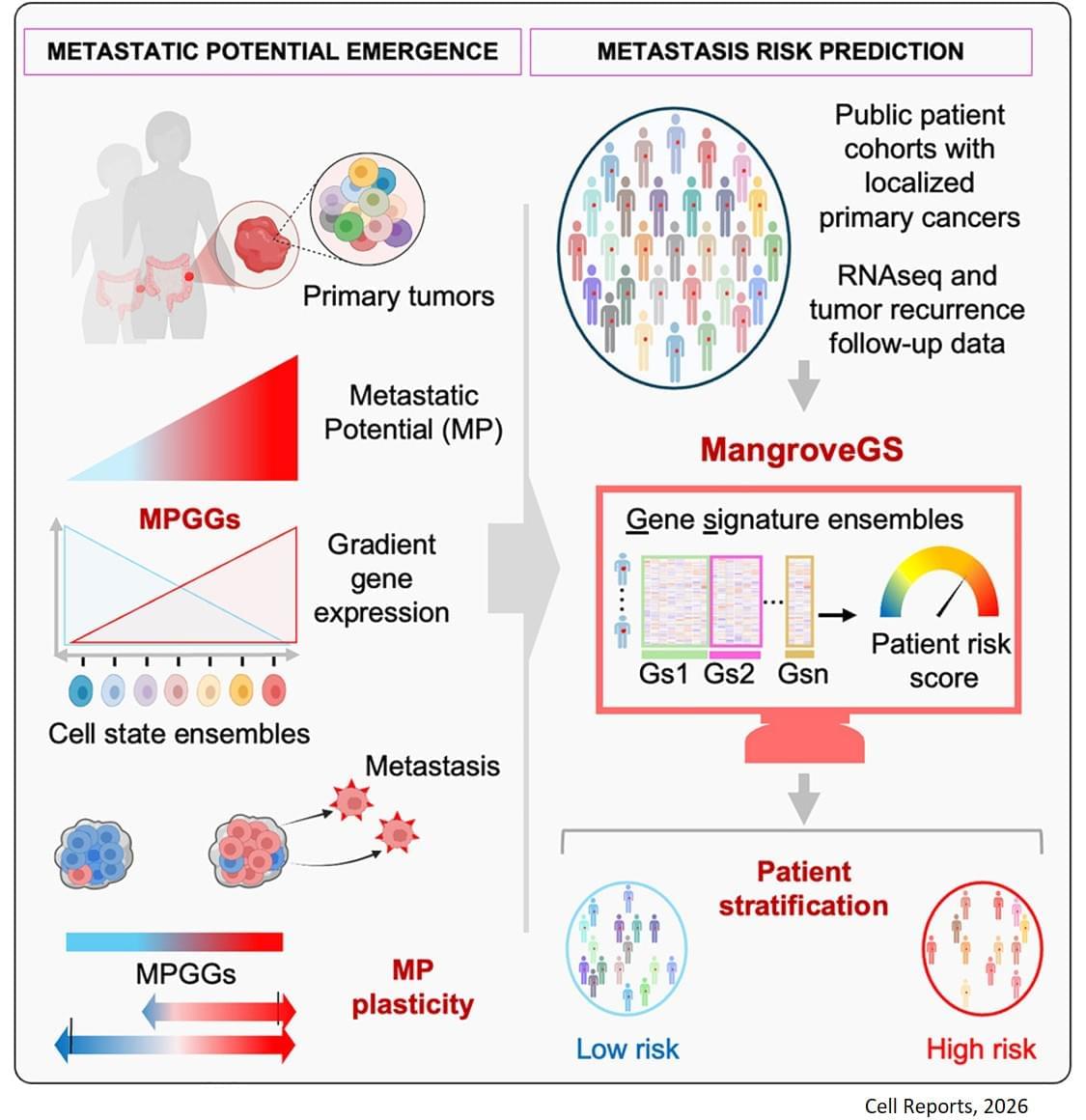
Metastasis remains the leading cause of death in most cancers, particularly colon, breast and lung cancer. Currently, the first detectable sign of the metastatic process is the presence of circulating tumor cells in the blood or in the lymphatic system. By then, it is already too late to prevent their spread. Furthermore, while the mutations that lead to the formation of the original tumors are well understood, no single genetic alteration can explain why, in general, some cells migrate and others do not.
“The difficulty lies in being able to determine the complete molecular identity of a cell – an analysis that destroys it – while observing its function, which requires it to remain alive,” explains the senior author. “To this end, we isolated, cloned and cultured tumor cells,” adds a co-first author of the study. “These clones were then evaluated in vitro and in a mouse model to observe their ability to migrate through a real biological filter and generate metastases.”
The analysis of the expression of several hundred genes, carried out on about thirty clones from two primary colon tumors, identified gene expression gradients closely linked to their migratory potential. In this context, accurate assessment of metastatic potential does not depend on the profile of a single cell, but on the sum of interactions between related cancer cells that form a group.
The gene expression signatures obtained were integrated into an artificial intelligence model developed by the team. “The great novelty of our tool, called ‘Mangrove Gene Signatures (MangroveGS)’, is that it exploits dozens, even hundreds, of gene signatures. This makes it particularly resistant to individual variations,” explains another co-first author of the study. After training, the model achieved an accuracy of nearly 80% in predicting the occurrence of metastases and recurrence of colon cancer, a result far superior to existing tools. In addition, signatures derived from colon cancer can also predict the metastatic potential of other cancers, such as stomach, lung and breast cancer.
After training, the model achieved an accuracy of nearly 80% in predicting the occurrence of metastases and recurrence of colon cancer, a result far superior to existing tools. In addition, signatures derived from colon cancer can also predict the metastatic potential of other cancers, such as stomach, lung and breast cancer.
Thanks to MangroveGS, tumor samples are sufficient: cells can be analysed and their RNA sequenced at the hospital, then the metastatic risk score quickly transmitted to oncologists and patients via an encrypted Mangrove portal that has analysed the anonymised data.
“This information will prevent the overtreatment of low-risk patients, thereby limiting side effects and unnecessary costs, while intensifying the monitoring and treatment of those at high risk,” adds the senior author. “It also offers the possibility of optimising the selection of participants in clinical trials, reducing the number of volunteers required, increasing the statistical power of studies, and providing therapeutic benefits to the patients who need it most.” ScienceMission sciencenewshighlights.

In Ruijs-Aalfs progeria syndrome, patients experience accelerated aging and liver cancer.
Now, scientists showed that mutations in a certain gene prevent cells from repairing DNA damage during mitosis, triggering inflammatory immune responses that may fuel premature aging.
Read more.
Researchers have shown that harmful bonds between protein and DNA fuel immune attack in progeria. Pumping up a protein that cuts these bonds could prevent symptoms.
Procrastination, the tendency to unnecessarily delay or put off tasks even if this will have negative consequences, is a common behavior for many people. While occasionally delaying or putting off bothersome tasks is not necessarily problematic, severe and prolonged procrastination is closely tied to some neuropsychiatric disorders, including attention-deficit/hyperactivity disorder (ADHD) and anxiety disorders.
Unveiling patterns in the brain’s structure and genetic factors linked to procrastination could help to reliably uncover this tendency to postpone tasks in affected individuals. This could in turn inform the development of preventative strategies or interventions that tackle procrastination early, before it exacerbates other underlying mental health disorders.
Researchers at the Chinese Academy of Sciences and other institutes in China recently carried out a study aimed at shedding new light on the biological and genetic roots of procrastination. Their paper, published in Molecular Psychiatry, outlines specific patterns in the brain’s structure during adolescence that are linked to procrastination in adulthood.


The shared pathways were linked to neuron maturation, synapse formation, and the control of gene activity. Further analysis pointed to a group of genes involved in organizing DNA and regulating which genes are switched on or off. These genes sit high in the regulatory chain, influencing many downstream processes previously linked to autism.
To test whether this network played an active role, the team reduced the activity of several key regulators using CRISPR-based methods in neural cells. This led to downstream changes similar to those seen in the autism models.
However, organoids from individuals with idiopathic autism showed less consistent changes, likely reflecting the complex and distributed genetic risk seen in most autism cases.
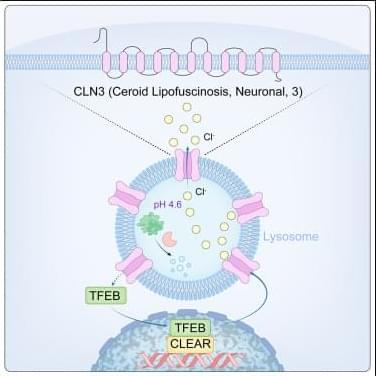
Lysosomes degrade damaged organelles and macromolecules to recycle nutrient components. Lysosomal storage diseases (LSDs) are linked to mutations of genes encoding lysosomal proteins and may lead to age-related disorders, including neurodegenerative diseases. But, how lysosomal dysfunction contributes to neurodegenerative diseases is not clear yet…
The researchers identify CLN3 (ceroid lipofuscinosis, neuronal 3), linked to Batten disease as a conserved lysosomal protein that regulates lysosomal chloride homeostasis, pH, and protein degradation.
Curcumin analog C1 is a natural compound with anti-inflammatory properties could enhance CLN3 activity and improve lysosomal function by activating TFEB. sciencenewshighlights ScienceMission https://sciencemission.com/CLN3-n-chloride-efflux-n-lysosomes
Wang et al. identify CLN3 as a conserved lysosomal protein that regulates lysosomal chloride homeostasis, pH, and protein degradation. Transcription factor EB (TFEB) activation enhances CLN3 function, revealing the TFEB-CLN3 signaling axis as a promising therapeutic target for lysosomal storage disorders.