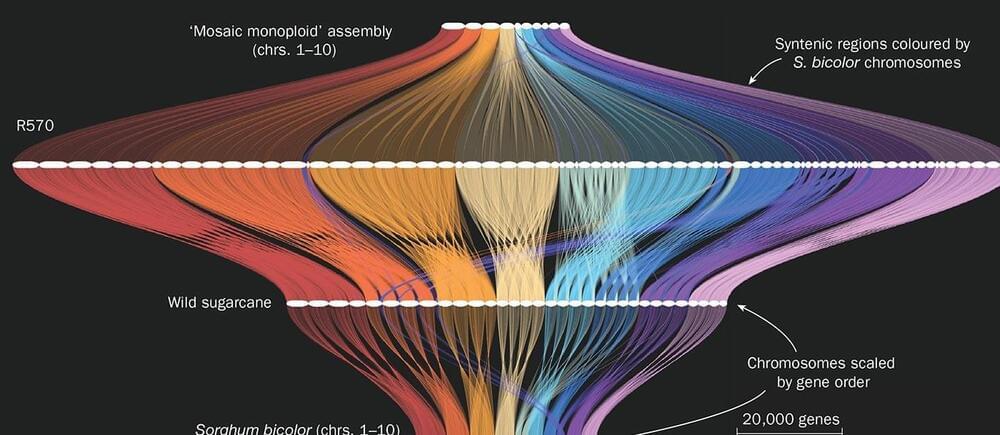
“The increases in THC levels found in cannabis could mimic some of the more pronounced effects that we see for people who are slower metabolizers,” said Dr. Christal Davis.
How can genetics influence cannabis consumption? This is what a recent study published in Addictive Behaviors hopes to address as a team of researchers investigated a link between how genetic variances influence how a person metabolizes THC, which could not only determine future use but also the chances of succumbing to cannabis use disorder, or CUD. This study holds the potential to help cannabis users, medical professionals, legislators, and the public better understand the physiological influences of cannabis use, even at the molecular level.
For the study, the researchers enlisted 54 participants between 18–25 years of age, 38 of whom suffered from CUD while the remaining 16 suffered from non-CUD substance abuse. It has been determined that individuals aged 18–25 have a three times greater likelihood of having CUD compared to individuals over the age of 26. After obtaining blood samples from each participant, the researchers tested them for differences in gene markers, specifically pertaining to THC-metabolizing enzymes. Additionally, each participant was instructed to fill out a questionnaire regarding their experiences with cannabis use and how it makes them feel when they use it.
In the end, the researchers found notable differences between men and women participants, specifically regarding how young women with CUD were found to metabolize THC at slower rates than young women who did not suffer from CUD. For the men, the researchers discovered negative reports from cannabis use with those who also metabolized THC at slower rates, which was the same for both sexes. Additionally, the researchers’ found CUD was more prevalent in individuals who started using cannabis when teenagers, as well. The researchers concluded that proper treatment options for CUD could be proposed due to lack of genetic testing.