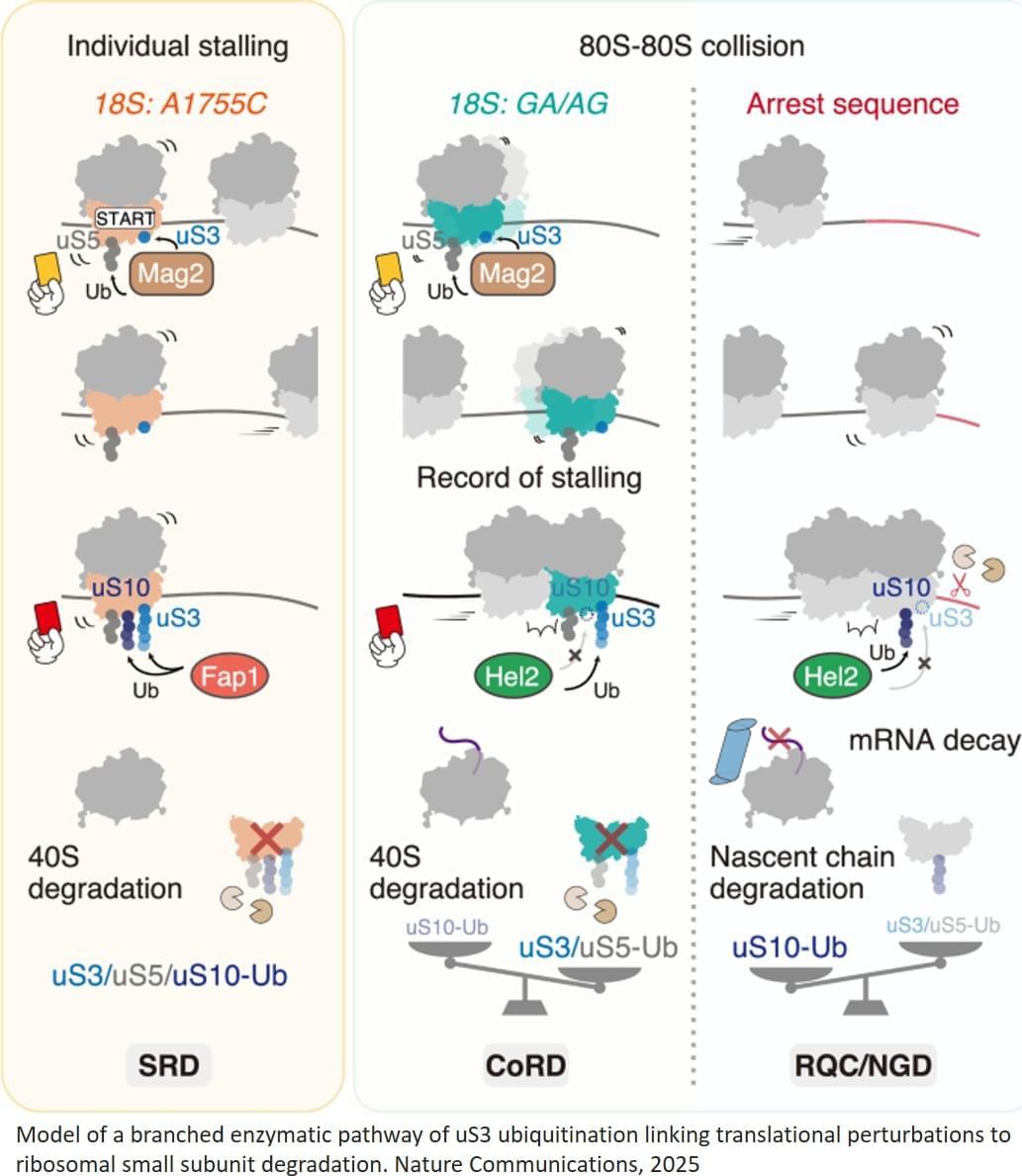
The androids of the future will be the distant results of synthetic biology and not silicon.
🚀 Step into a world of boundless innovation as we take you on a journey through the awe-inspiring technologies that await humanity in the 22nd century! 🌌 From advancements in space exploration to mind-boggling leaps in artificial intelligence, this captivating video offers a glimpse into the cutting-edge breakthroughs that will redefine the very fabric of human existence.
🌐 Witness the birth of extraterrestrial civilizations as humans venture further into space, exploring distant planets and establishing self-sustaining colonies. Experience the seamless integration of artificial intelligence into our daily lives, transforming how we interact with technology and creating new possibilities for societal progress. Prepare to be amazed by quantum computing’s extraordinary power, revolutionizing problem-solving and opening doors to scientific discoveries previously deemed impossible.
🌿 Delve into the world of sustainable marvels, where eco-friendly innovations mend our relationship with the environment and pave the way for a greener, more harmonious future. Explore the ethical implications of biotechnology advancements, which offer insights into longevity and human potential. This video paints an inspiring picture of the limitless possibilities and profound transformations that lie ahead in the remarkable world of 22nd-century technologies. Like, share, and subscribe to our channel for more captivating glimpses of the ever-evolving world of tomorrow. 🌟🔮🌠 #FutureTechnologies #22ndCenturyInnovations #EmbracingTomorrow