Researchers at Kanazawa University report in ACS Nano how high-speed atomic force microscopy can be used to study the biomolecular mechanisms underlying gene editing.
The DNA of prokaryotes—single-cell organisms, for example bacteria—is known to contain sequences that are derived from DNA fragments of viruses that infected the prokaryote earlier. These sequences, collectively referred to as CRISPR, for “clustered regularly interspaced short palindromic repeats,” play a major role in the antiviral defense system of bacteria, as they enable the recognition and subsequent neutralization of infecting viruses. The latter is done through the enzyme Cas9 (“CRISPR-associated protein 9”), a biomolecule that can locally unwind DNA, check for the existence of the CRISPR sequence and, when found, cut the DNA.
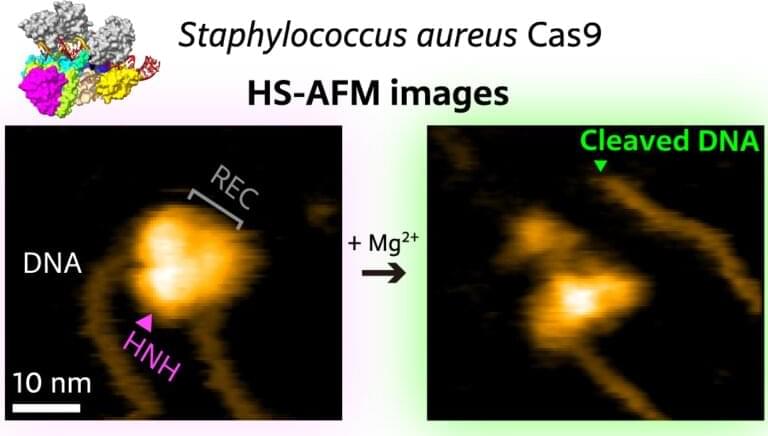
In recent years, CRISPR/Cas9 has emerged as a genome editing tool based on the notion that the Cas9 protein can be activated with artificially created CRISPR-like sequences. Sometimes, however, the wrong target is “caught” by Cas9—when the wrongly identified DNA sequence is too similar to the intended target sequence. It is therefore of crucial importance to fully understand how Cas9 binds to, “interrogates,” and cuts DNA. Mikihiro Shibata from Kanazawa University and colleagues have now succeeded in video-recording the DNA binding and cleaving dynamics of Staphylococcus aureus (a particular bacterium) Cas9 by means of high-speed atomic force microscopy (HS-AFM). Their observations will help to reach a more complete understanding of CRISPR-Cas9 mechanisms.









Comments are closed.